The main goal, and challenge, in single-cell RNA sequencing data analysis is assigning cell type identity to cells. Performed after dimensionality reduction and clustering, this step gives a name to each cluster of cells. Even though annotation remains a difficult feat, we have tools and methods that attempt to solve the problem. In this blog, we will summarize some common approaches along with their pros and cons.
What is defined as cell type?
Cells are the basic building blocks of tissues and organisms, carrying out highly specific and specialized functions. Cells specialize as a result of various intrinsic (molecular profiles) and extrinsic (spatial location) cues. Humans naturally want to classify everything into little boxes; thus we should ask ourselves “What exactly is cell type?”. Despite research efforts over the past 3-4 centuries, there is still a lot more to learn about cells.
The annotation question is largely about specifying the levels of analysis.
The annotation question is largely about specifying the levels of analysis. What level are we classifying at? Functional? Regulatory? Morphological? How much variation are we expecting? Gene expression levels vary gradually over a spectrum, and without having a level of analysis in mind, we cannot draw boundaries between different expression “types”. More importantly, transcriptional differences that we use as separation boundaries may not have any biological functional relevance at all.
Moreover, if we take into consideration cell development and fate decisions, the question is even more challenging. The changes of cells throughout their life cycles are intricate and rapid. Assume that we could rigorously classify cell types and states in our desired level of analysis, we still need to make sense of such dynamic biological regulation.
Over the past decade or so, single-cell sequencing technologies have allowed us to zoom into the molecular mechanisms that regulate cell behavior, including fate decisions, developmental transitions, and responses to injuries and diseases. Thanks to the technology, we can conduct mixed cell experiments without experimentally purifying or separating the cells by type. We then computationally utilize unbiased algorithms to label cell types in multiple species, tissues, and contexts, and also construct systems biology models to predict cell behaviors during development.
What approach should we use?
We can broadly classify single-cell cell type annotation into two classes: manual and automated. In manual methods, we use marker features to define cell identities, one by one. In automated methods, we use computational tools to analyze the transcriptomic profiles and assign identities. Each option has its pros and cons.
Manual tools
Manual annotation is straightforward – through marker genes. You can do so in a “forward” fashion, by using a list of marker genes as reference, then query and match reference with selected cells.
We can do this within Seurat with Marker Features function, or by Differential Expression Analysis (DEA) of a cell population against others.
We can often find marker genes in the single-cell atlas, especially if our sample comes from a similar biological context as the ones used in the atlas. Researchers are also recommended to mine the literature for further evidence. In the best-case scenario, if most cells in our cell population show high expression levels of canonical markers, then the annotation is almost complete.
We know too well that such a happy scenario is hardly a reality. There are times when the classic marker genes are insufficient for classification, where we meet a novel or rare cell type or, the cells were poorly handled during the experiment. If we are fortunate to have avoided these issues, we may proceed with enrichment analysis to find out the cell functions.
Alternatively, you can use machine learning to crawl for similar cell populations within databases for single-cell studies. This method, though more advanced, will return not only the suggested cell type but also suggest similar signature genes and processes. These suggestions, akin to Google search suggestions, may help you dive deeper into your studies. And with that, let’s explore some automated tools.
Automated tools
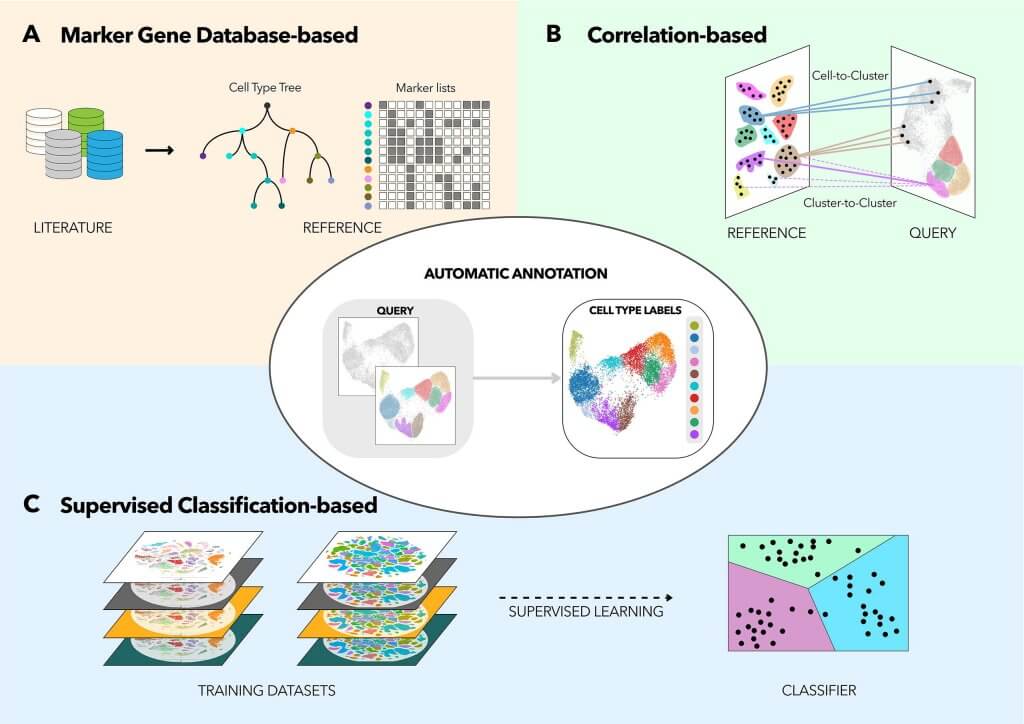
Automated methods are more diverse than manual methods, and are based on one of three core principles:
- Marker-based: algorithms score, and rank clusters based on the level of expression of marker gene, then assign clusters to their respective cell type. Some open-source tools include SCINA , CellAssign.
- Supervised classification: we use machine learning to train the tools with reference single-cell studies. For example, we have: scClassify and scPred. More training datasets and better quality training datasets will result in higher-quality tools.
- Correlation-based: As the name suggests, we calculate how much each cluster correlates to the reference cell type. We will also rank all the correlation scores and assign clusters to whichever cell type with the best score. The most familiar example is: Seurat.
Final note
Tools are created to answer the problem that their creators have been facing, and none of them are perfect. Depending on the scale and the type of your dataset, some methods may be better than others. You must always follow up and validate your cell annotation. You can test the statistical significance of cluster membership, and also integrate cell annotation with other experimental and computational methods. The best practice is to use one or two automated tool(s) to quickly give your cells some identities, and then cross-check with a manual method and other validation methods.
If you need scRNA-seq-related help, Dolomite Bio offers end-to-end Single-Cell Consultancy Service (SCC) and Bioinformatics Service (BIS) that helps you through one or more steps of the workflow:
Sample preparation (SCC)
Library preparation (SCC)
Sequencing (SCC)
Computational Analysis (SCC + BIS)
Interested queries and/or suggestions for what we should write next in our blog series should be directed to: bioinformatics@dolomite-bio.com
References
- Trapnell, C. (2015) Defining cell types and states with single-cell genomics– Genome Research.
- Pasquini, G., Arias, J., Schafer, P., Busskamp, V. (2021) Automated methods for cell type annotation on scRNA-seq data – Computational and Structural Biotechnology Journal 19.
- Clarke, Z., Andrews, T., Atif, J., Pouyabahar, D., Innes, B., MacParland, S., Bader, G. (2021) Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods . Nature Protocols 16, 2749-2764.
- Amezquita, R., Lun, A., Hicks, S., Gottardo, R. (2021) Basics of Single-Cell Analysis with Bioconductor . Bioconductor, 2021.
- Nguyen, H. (2022) scRNA-Seq Cell Type Annotation: Common Approaches and Tools. BioTuring.
- Ianevski, A., Giri, A., Aittokallio, T. (2022) Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data]. Nature Communications 13.
Check out the rest of our Bioinformatics blogs
Pre-processing of single cell data – A comparison of dropSeqPipe and Kallisto-bustools