T helper (Th) cells play a central role in immunity, an avidly discussed topic in today’s applied and basic research environments. Th17 cells are key regulators of inflammation and undergo significant metabolic changes following activation to initiate an effective inflammatory response. However, the epigenetic modifications regulating T cell metabolism remain largely unknown despite their crucial role in the human immune response.
In a recent publication, Cribbs et al. established an epigenetic link between the histone H3K27me3 demethylases KDM6A/B and the coordination of a metabolic response in T cells [1].

As part of this extensive study, the authors show that the histone H3K27 demethylases KDM6A and KDM6B are critical regulators of Th17 pro-inflammatory function in humans. Specifically, by using droplet based single-cell sequencing (scRNA-Seq) they were able to demonstrate that treatment with GSK-J4, a KDM6 inhibitor, impacts on Th17 function and leads to reduced metabolic activity. The powerful analytical genomic techniques showcased in the paper have allowed researchers to better understand the functional consequences arising from inhibiting KDM6A/B in sub-populations of pro-inflammatory cells.
But how can modern day sequencing techniques delve into such complex cell type interactions with the latest instrumentation? And importantly, how can these valuable conclusions contribute to future treatment development within the fast-paced field of immunology?
scRNA-Seq: Why one cell can give more information than many
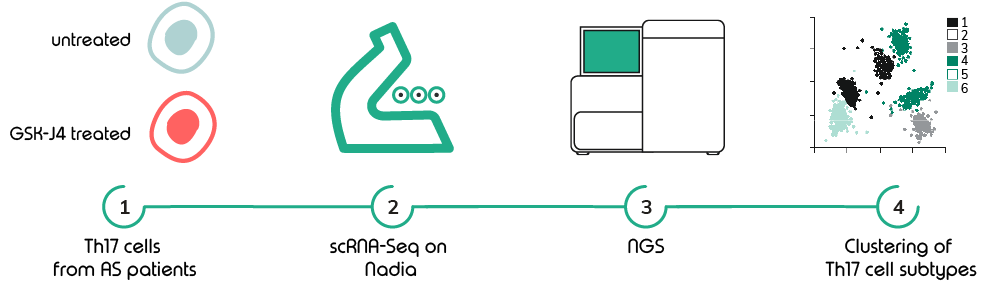
Using the Nadia Instrument (Dolomite Bio), a droplet-based microfluidic platform for running standardised automated single cell protocols, the authors performed single-cell RNA sequencing on Th17 cells isolated from patients suffering the inflammatory condition Ankylosing Spondylarthritis (AS). Prior to encapsulation, the cells were treated with histone deacetylase inhibitors of KDM6A/B and their anti-inflammatory effects observed at the single-cell level. Using this potent technique, changes in transcriptomic responses and metabolic phenotypes were evaluated at single cell resolution.
Whilst bulk RNA sequencing can be used to draw conclusions about generalised transcript changes within a tissue, this represents an average of all the transcriptional changes from each cell. Single-cell technology has allowed an understanding of the inter-individual heterogeneity with respect to disease and treatment outcomes, which would not have been possible with traditional bulk transcriptomic techniques. Therefore, technology platforms such as the Nadia Instrument are critical for driving novel and informative biology.
Very few tissues are truly homogeneous in nature, instead being composed of a multitude of cell types. Even for single cell type, such as the T cells examined in this study, precise cellular identity exists on a continuum, where the variation between individual cell transcriptomic states can be massive.
Cribbs et al. initially used bulk RNA-Seq data to detect the general transcriptional changes that occur following KDM6A/B inhibition, confirming previous hypotheses suggesting that KDM6A/B inhibition led to a general anti-inflammatory response. With the added resolution granted by scRNA-Seq however, the authors observed significant heterogeneity in the Th17 response to GSK-J4, despite those cells being isolated from a relatively homogeneous population. In this case, like in many other cutting-edge immunology research scenarios, single cell sequencing technologies such as those delivered by Dolomite Bio’s Nadia Instrument have proven invaluable for giving much needed context to bulk RNA-seq findings. With such a detailed methodology at hand, researchers like Dr. Adam Cribbs and his co-authors have begun to delve into the intricacies of human inflammatory disease in increasingly elegant ways to highlight valuable potential avenues of treatment.
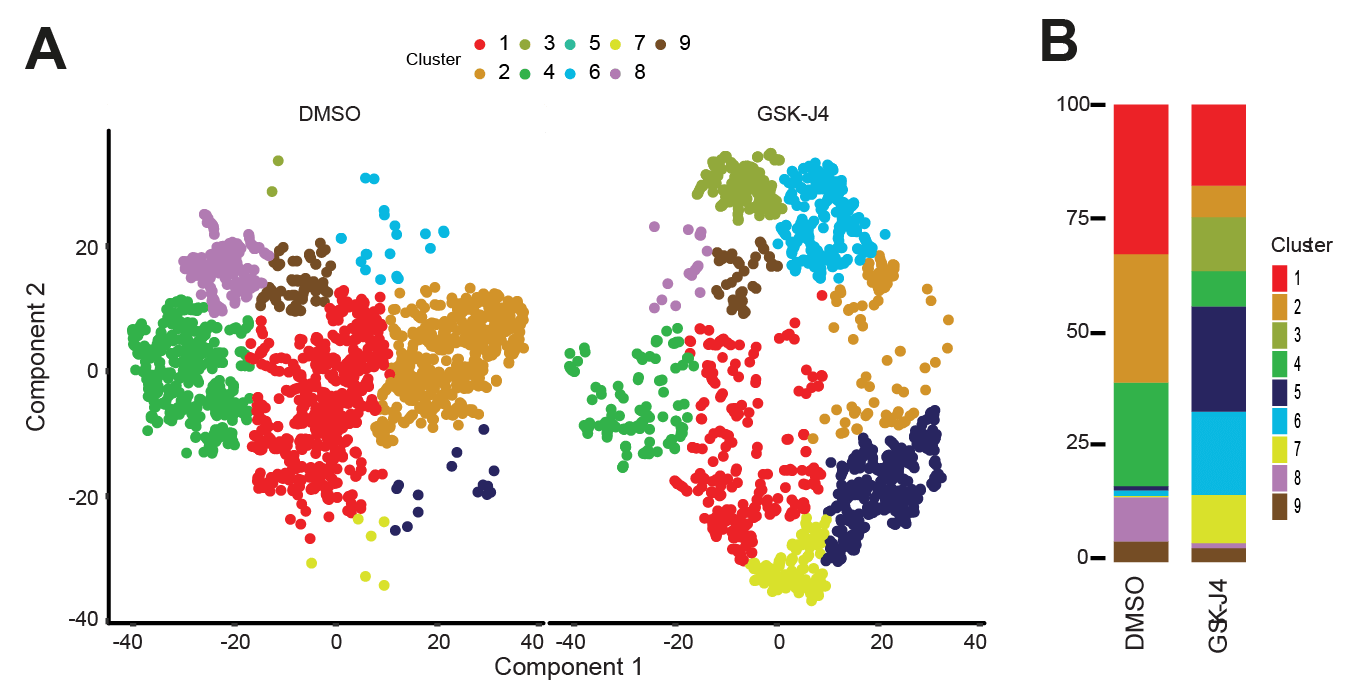
Single cell RNA sequencing elucidates a complex T cell response to KDM6A/B inhibition. (A) tSNE plots showing clustering of CD4+ T cells isolated from AS patients and enriched for Th17 cells with (right) and without (left) GSK-J4 treatment. (B) Proportional representation of each cell type cluster from each tSNE plot. Adapted from Figures 4A and 4B from Cribbs et al., 2020 with permission from authors.
Picking the right instrument for the job
Single cell RNA-Sequencing (scRNA-Seq) can often give us greater insight into the real biological picture than bulk RNA sequencing. However, to gain scRNA-Seq data for more than a few hundred transcriptomes, researchers must turn to the latest technology to tackle their research questions with sufficient throughput.
Using microfluidics to encapsulate single cells within picolitre sized droplets alongside oligo-coated capture beads [2], the Nadia platform allows researchers to easily and reproducibly generate libraries for thousands of single cell transcriptomes in a single run.

Despite this impressive throughput, users of the Nadia instrument need not sacrifice data quality. Presented by Cribbs et al., these datasets yield a median number of 2,835 genes per cell for several thousands of purified Th17 cells, ample datapoints to draw complex conclusions about single cell immune phenotypes, metabolism and cytokine function.
Thanks to this complex information provided by methodologies like single cell RNA-Sequencing, KDM6 enzyme inhibitors have been revealed as an interesting therapeutic strategy in autoimmune disease. But the fascinating methodologies used by Cribbs et al. don’t stop at scRNA-Seq. Read more about the epigenetic aspects of human Th17 cell development in Cribbs et al. (2020).
Interested in single cell RNA-Sequencing in immune cells or many other diverse cell types? Learn more about the Nadia Instrument here!
References
[1] Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proceedings of the National Academy of Sciences 2020. https://doi.org/10.1073/PNAS.1919893117.
[2] Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015;161:1202–14. https://doi.org/10.1016/j.cell.2015.05.002.