The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the rise of the coronavirus disease 2019 (COVID-19) pandemic with over 10 million confirmed cases worldwide at the time of writing. Since then, a plethora of literature has been published around COVID-19 trying to understand the viral replication, host responses and disease progression.
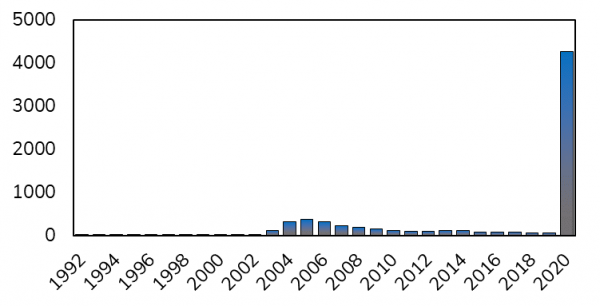
Timeline of publications containing the keywork SARS-CoV since 1992. With spikes after the outbreak of ‘old’ SARS-CoV in 2002/2003 and now in 2020 during the outbreak of SARS-CoV-2.
In contrast to 2002/2003, during the outbreak of the SARS-CoV virus, we now have a new tool with single cell RNA-Sequencing that allows us to understand the heterogeneity of viral infections at an unprecedented level. As Emanuel Wyler from the Max Delbrück Center (MDC) in Berlin says: “Virus infections are a very heterogeneous process. Already in a simple cell line model, the amount of virus particles entering the cells and the progression of infection can vary a lot. Methods averaging many cells such as bulk RNA-seq would mask these differences.” (Read the full interview here)
A pre-print recently published by Emanuel Wyler a researcher in the Landthaler group at the MDC for molecular medicine in Berlin uses bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines to identify molecular targets for therapeutic intervention (1). Their study compared the immune response of human cell lines (H1299, Caco-2 and Calu-3 cells) to the “old” SARS-CoV virus from 2002/2003 and to the “new” SARS-CoV-2 from 2019.
This study led to 4 main findings:
- ScRNA-Seq shows SARS-CoV-2 leads to a much stronger immune response than SARS-CoV
- Virus infected cells showed induction of the microRNA miR-155
- RNA velocity identified the sequential induction of IRF and the NF-kappaB signalling pathways
- Inhibition of Hsp90 with the known cancer drug 17-AAG lead to reduced viral growth
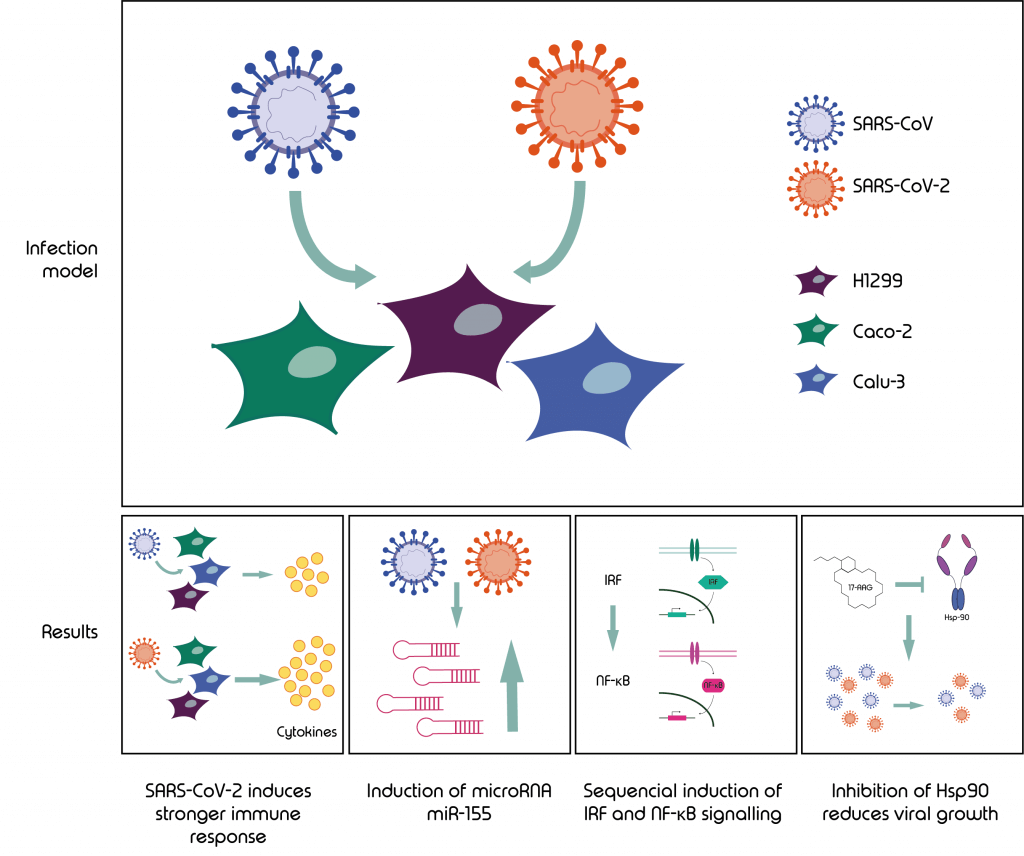
Graphical abstract: This study lead to four main finings using a model of SARS-CoV and -2 infected cell lines H1299, Caco-2 and Calu-3.
We will now dive deeper into the previously mentioned points and explain step by step the findings in detail. However, we will first briefly discuss the differences observed in the cell lines used as infection models.
Differences observed in SARS-CoV-2 infected cell lines
Virus replication and host cell responses were studied in the epithelial lung cancer cell lines, H1299 and Calu-3, and the epithelial colorectal adenocarcinoma cell line, Caco-2. The latter is often used as a coronavirus cell culture model and was already employed for other SARS-CoV and SARS-CoV-2 comparative studies (2,3). Expression of IFIT1, IFIT2 and OAS2 in H1299 and Calu-3 indicated that the sensing of cytoplasmic foreign RNA is active in these cell types, which was not observed in Caco-2 cells. The percentage of viral transcripts in the cellular RNA was lowest in H1299 cells. The authors speculated that this might be partially due to the low expression of Angiotensin-Converting Enzyme 2 (ACE2), a SARS-CoV receptorACE2 is the entry receptor utilized by SARS-CoV and SARS-CoV-2 to mediate infection. Transcriptional activation of this receptor has been seen in SARS-CoV-2 infected cells (4) and has also been confirmed in the publication discussed in this review. Interestingly, another publication showed that treating a cell line known to permit SARS-CoV replication with an Anti-ACE-2 Antibody disrupted the interaction between virus and receptor (5).
ScRNA-Seq shows SARS-CoV-2 leads to a much stronger immune response than SARS-CoV
The authors used the Nadia Instrument to perform scRNA-Seq on Calu-3 cells at different time points post infection with both, SARS-CoV and SARS-CoV-2. After 4h the percentage of cells with detectable amounts of viral RNA were between 40 and 60% which increased to 100% after 8 hpi (hours post infection) with viral load being comparable between the two viruses.
One of the findings with possible clinical relevance was that SARS-CoV-2 infected Calu-3 cells showed a two-fold higher induction of interferon stimulated genes (ISGs) compared to cells infected with SARS-CoV infections. This is interesting as cytokines such as CXCL10 or IL6 are induced. This in turn can lead to the recruitment of an abnormally high number of immune cells which was seen to cause lung damage in COVID-19 patients, due to what’s called an acute respiratory distress syndrome (ARDS) (6).
What they found in detail was that in bulk as well as in scRNA-Seq data the expression of ISGs, IFIT1 and IFIT2 was particularly strong in SARS-CoV-2 infected cells. Furthermore, for IFIT2, the result indicated a likely off-on switch in SARS-CoV-2 infections between 4 hpi and 8 hpi, with the expression levels being independent of the amount of viral RNA. This switch was not observed for SARS-CoV. Notably, cells infected with SARS-CoV-2 showed stronger alterations of their transcriptome than cells infected with the “old” SARS virus.
Virus infected cells show induction of the microRNA miR-155
Small RNA profiling of Calu-3 cells has shown strong upregulation of miR-155-3p and -5p, originating from the host gene miR-155, which was also highly expressed in infections with both viruses. MiR-155 microRNAs are associated with immunity and inflammation and have been connected to a variety of viral infections. Interestingly, in a study with mice infected with influenza A virus, lung damage through ARDS could be reduced by deletion of miR-155, making this microRNA a potential therapeutic target for COVID-19 patients (7).
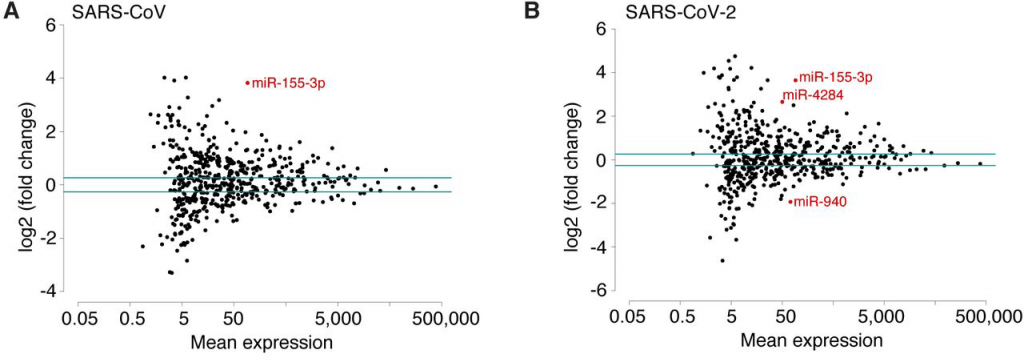
Scatterplots of miRNA mean expression comparing SARS-CoV and SARS-CoV-2 and indicating in red differentially expressed miRNA.
RNA velocity identifies sequential induction of the IRF and the NF-kappaB signalling pathways
The authors used a computational method published in 2018 called RNA velocity (8). The method aims to help with the analysis of time-resolved phenomena by distinguishing between un-spliced and spliced mRNAs in scRNA-Seq protocols and thereby predicts the future state of individual cells. This analysis suggested that during infection of Calu-3 cells, the induction of interferons is short, with IFIT2, IFIT1 or OAS2 showing high intron counts during accumulation of viral RNA. When the signal for interferon regulatory factors (IRFs) decreases, the intron counts for NF-κB target genes are still high. This suggests that IRFs activities precede that of NF-κB and that both only overlap during a short period of time.
Inhibition of Hsp90 with the known cancer drug 17-AAG leads to reduced viral growth
To identify changes in the cellular transcriptome that are not connected to the interferon response the authors performed scRNA-Seq on H1299 cells which are less susceptible to the viral infection. They noticed a higher expression of HSP90AA1, transcript of Heat shock protein 90 (HSP90), in cells with SARS-CoV-2 viral RNA compared to those without, which was also observed in Calu-3 cells. HSP90 is a molecular chaperone and a protein of relevance in the infection of a range of viruses (9). This chaperon can be inhibited by Tanespimycin/17-N-allylamino-17-demethoxygeldanamycin (17-AAG), a derivative of the antibiotic geldanamycin (10). Although the treatment with 17-AAG showed no apparent effect on cell viability, the authors were able to detect an inhibition of viral replication by 50% at high nanomolar concentrations (500-800 nM).
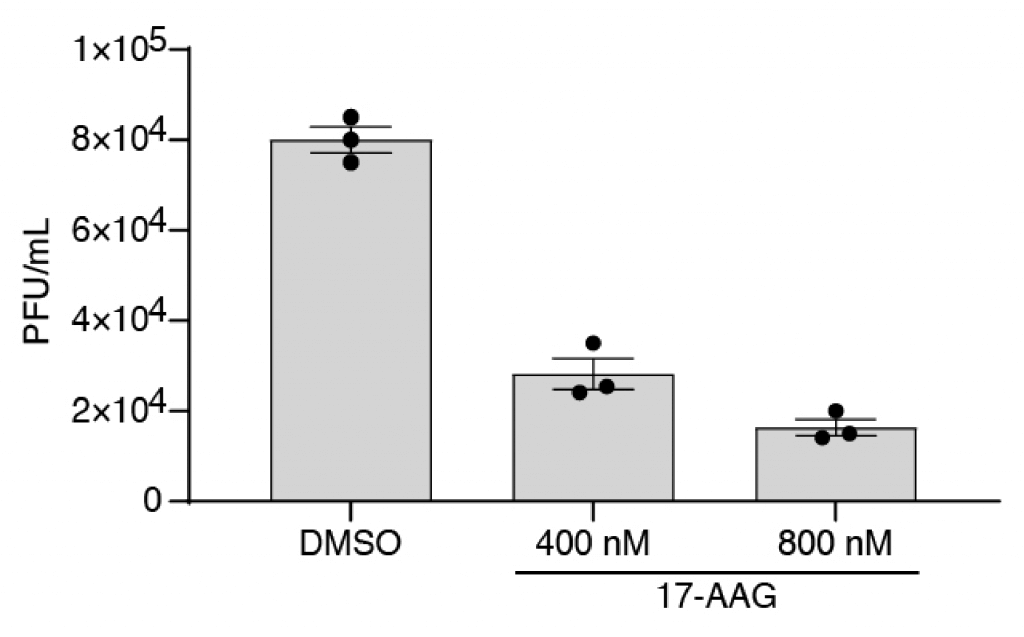
Reduced viral growth through inhibition of Hsp90 with 17-AAG
On a transcriptional level the authors observed reduced expression of INFL1, IL1B and TNF which were not observed to that level for IFIT2. Which in turn supports the finding of a “on-off-switch” independent of the amount of viral RNA observed.
Summary
This study was able to identify two interesting targets of potential clinical relevance. The deletion of microRNA miR-155 led to reduced lung damage caused by ARDS and the inhibition of HSP90, a molecular chaperone, leading to the reduction of viral replication.
The latter could pose a promising target, as several inhibitors are already in phase 2 and 3 clinical trials as anticancer drugs and are therefore readily available as therapeutics for COVID-19 patients. However, the extent to which the observations shown on cell lines in this study are reproducible in animal models and humans are unknown and will require further investigation.
Further reading
Read our interview with Dr Emmanuel Wyler, discussing this paper:
Or you can read our blog looking in depth at immunology research on the Nadia Instrument:
scRNA-Seq in Immunology – A Case Study of the Nadia instrument
References
- Emanuel W, Kirstin M, Vedran F, Asija D, Theresa GL, Roberto A, et al. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. bioRxiv. 2020 May 5;2020.05.05.079194.
- Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020 May 1;1(1):e14–23.
- Bojkova D, McGreig JE, McLaughlin K-M, Masterson SG, Widera M, Krähling V, et al. SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv. 2020 Apr 5;2020.04.03.024257.
- Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell [Internet]. 2020 Apr 27 [cited 2020 May 27]; Available from: http://www.sciencedirect.com/science/article/pii/S0092867420305006
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 Apr 16;181(2):271-280.e8.
- Zhang C, Wu Z, Li J-W, Zhao H, Wang G-Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. 2020 May;55(5):105954.
- Woods PS, Doolittle LM, Rosas LE, Nana-Sinkam SP, Tili E, Davis IC. Increased expression of microRNA-155-5p by alveolar type II cells contributes to development of lethal ARDS in H1N1 influenza A virus-infected mice. Virology. 2020 Jun 1;545:40–52.
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018 Aug;560(7719):494–8.
- Geller R, Taguwa S, Frydman J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta. 2012 Mar;1823(3):698–706.
- Dimopoulos M-A, Mitsiades CS, Anderson KC, Richardson PG. Tanespimycin as antitumor therapy. Clin Lymphoma Myeloma Leuk. 2011 Feb;11(1):17–22.


