- Low doublet rates – Sample stirring to avoid settling or clumping of samples.
- Broad sample compatibility –Suitability for a wide range of tissue types and organisms.
- Application versatility –Option for protocol development.
- Exceptional data yield – High gene capture rate even at low sequencing depths.
Droplet-based microfluidic single cell platforms represent the pinnacle of single cell research technology today. Their ability to compartmentalise cells with unprecedented throughput ready for diverse applications is shaping the way biology is explored. As more biological fields move into the realm of single cell genomics, personalised medicine and high throughput cellular screening or analysis, there has never been a better time to consider droplet-based single cell technology.
But how can a researcher ensure that their equipment is both robust and future proof enough to meet potential advances in the rapidly flourishing field of single cell genomics? Researchers from clinical applications to basic research to applied industry should take care to consider these must-haves when equipping their labs for the single cell revolution.
Truly single cell
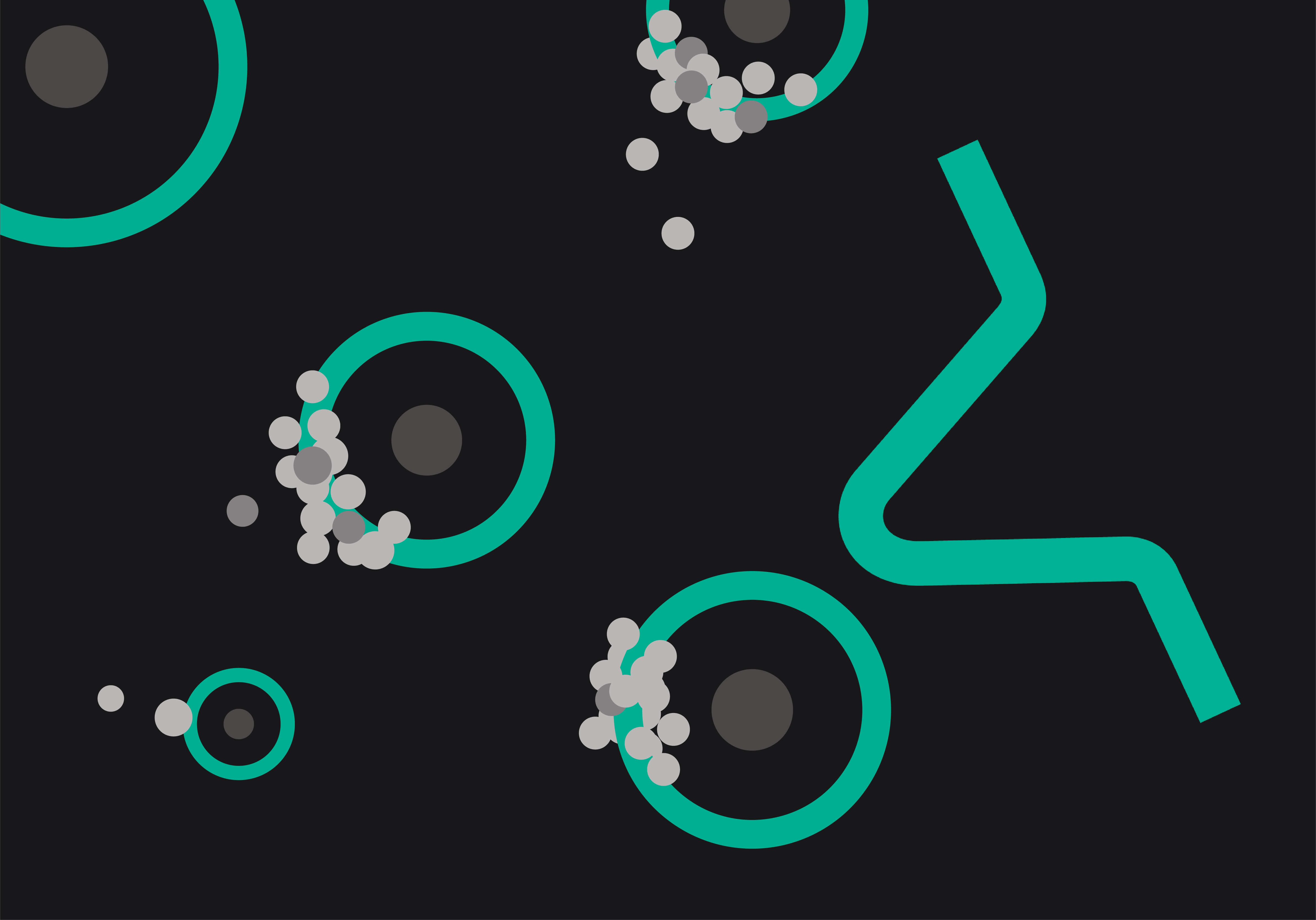
Simplistically, the central process within many single cell workflows is assigning cells and oligo capture beads into droplets [1]. This can be extremely efficient, but if it is not tightly controlled, multiple cells can be placed into the same droplet, creating doublets in the resulting data. High doublet rates are to be avoided. Why invest in a single cell platform if it doesn’t yield data from true single cells?
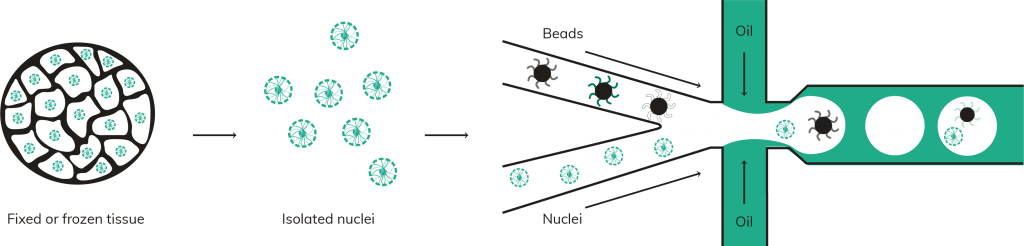
Whilst computational advances are being made to reduce the impact of doublets in single cell data [2–4], the most effective way to avoid doublets is to ensure that only single cells are partitioned into droplets to begin with. Many cell or sample types are inherently adhesive or dense, making them prone to agglutination or settling within the reservoirs of your single cell platform of choice. The true test of efficacy is not just how a single cell platform can partition standard cultured cell types, but whether it can retain low doublet rates even with difficult to separate sample types. Nuclei for example can be particularly problematic, often sticking together in simple buffers and inflating doublet counts.
Mechanical solutions are however available to reduce this issue. Uniquely among commercial platforms, the Nadia platform by Dolomite Bio ensures that cells and nuclei remain singulated for the entire duration of encapsulation with proprietary on-chip stirring of individual samples. By keeping sample solutions fully suspended for the entire encapsulation procedure, low doublet rates can be maintained even with tricky samples [5].
Broad sample compatibility
 Not all cells are created equal, even within a single organism or tissue. It is these differences
Not all cells are created equal, even within a single organism or tissue. It is these differences
that are often the target of investigation for single cell researchers [6].
Differences in morphology and physical size of cells can impact whether these cells can flow unhindered through a microfluidic system, potentially introducing biases in the resulting data.
So what hardware features does a microfluidic device need to make it compatible with any cell types that users might expect to study in the future?
Ability to alter buffer composition and droplet size can allow improved flexibility for cell types large and small, like bacteria or nuclei [5], and even cells that require specialised buffer conditions, such as plant protoplasts [7].
Illustrating this, cardiomyocytes are major cells of interest in prospective cell atlases, but their large size precludes them from being dropletised whole by some microfluidic systems. In some cases, researchers have compromised by processing single cardiomyocyte nuclei rather than analysing entire cell transcriptomes [8]. By contrast, single cell platforms like the Nadia instrument can encapsulate cells up to 60 µm in size, broadening the range of cell types accessible for transcriptomics.
As the blossoming world of single cell opens up to more and more model organisms, from plants to bacteria[9,10], and diverse cell types [6], the importance of single cell instrumentation which can accommodate varied cell types is more important than ever.
Experimental versatility
In the decade since single cell technologies first joined the genomics scene, both the number of cells able to be examined and the number of different methodologies has expanded rapidly [11,12]. For those with an ear to the ground in the field of Single Cell, it can sometimes seem that researchers develop a new -Seq every week. Any single cell platform worth its salt must therefore be sufficiently open to perform new and potentially unforeseen applications.
Whilst locked-in systems with a discrete set of applications are typically popular for their ergonomic design, users can be left behind when ground-breaking new molecular techniques surface. With a system capable of open protocol development and optimisation, such as the Nadia Innovate, researchers can proactively develop applications to take advantage of the latest in sequencing technologies.
Exceptional data yield
 In the single cell research field, advances in technology lead to improvements in data quality. Microfluidic hardware is no different. However, the relative quality of single cell data is measured based on what question is being asked. Some research can prioritise monodispersity or precision of droplet generation, such as using microfluidic equipment to place single cells into hydrogel droplets ready for growth studies or FACS (here).
In the single cell research field, advances in technology lead to improvements in data quality. Microfluidic hardware is no different. However, the relative quality of single cell data is measured based on what question is being asked. Some research can prioritise monodispersity or precision of droplet generation, such as using microfluidic equipment to place single cells into hydrogel droplets ready for growth studies or FACS (here).
Other applications like single cell RNA-Sequencing can prioritise sequencing metrics. Other than the previously described doublet rate, the main metric of interest is gene capture, a term for how many genes one can expect to glean from each single cell at a given sequencing read depth. When comparing datasets from different single cell platforms, it is important to standardise this data according to both sequencing depth per cell and throughput. Microfluidic devices excel at throughput, being able to assess hundreds of thousands of single cells. Due to these high cell numbers, individual sequencing depth per cell can be on the low side, so the best microfluidic single cell protocols need to yield high gene capture even at low sequencing depths.
Data quality is also affected heavily by cell type, so where possible comparisons should take place between like-for-like sample types. These kinds of comparisons within discrete fields are available in reviews for a variety of systems, from immune cytology [13,14] to crop plant research [10] to more generalised cell atlas projects [15].
Keeping all these metrics in mind, the best proof of a system’s efficacy with your cell type of choice is to test it yourself.
Contact Dolomite Bio today at applications@dolomite-bio.com to arrange a demonstration and find out whether the Nadia platform is the best fit for your research question.
Further reading
Read our previous blogs discussing Drop- Seq in greater depth
Drop-seq – Getting to the core of single cells one drop at a time
Or find our blog exploring immunology research with Dolomite Bio’s Nadia Instrument
scRNA-Seq in Immunology – A Case Study of the Nadia instrument
[1] Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015;161:1202–14. https://doi.org/10.1016/j.cell.2015.05.002. [2] Bais AS, Kostka D. Scds: Computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 2020;36:1150–8. https://doi.org/10.1093/bioinformatics/btz698. [3] DePasquale EAK, Schnell DJ, van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, et al. DoubletDecon: Deconvoluting Doublets from Single-Cell RNA-Sequencing Data. Cell Reports 2019;29:1718-1727.e8. https://doi.org/10.1016/j.celrep.2019.09.082. [4] Bernstein NJ, Fong NL, Lam I, Roy MA, Hendrickson DG, Kelley DR. Solo: Doublet Identification in Single-Cell RNA-Seq via Semi-Supervised Deep Learning. Cell Systems 2020;11:95-101.e5. https://doi.org/10.1016/j.cels.2020.05.010. [5] Davey K, Wong D, Konopacki F, Kwa E, Fiegler H, Sibley C. A flexible microfluidic system for single-cell transcriptome profiling elucidates phased transcriptional regulators of cell cycle. BioRxiv 2020:2020.01.06.895524. https://doi.org/10.1101/2020.01.06.895524. [6] Tang X, Huang Y, Lei J, Luo H, Zhu X. The single-cell sequencing: New developments and medical applications. Cell and Bioscience 2019;9:53. https://doi.org/10.1186/s13578-019-0314-y. [7] Davey MR, Anthony P, Power JB, Lowe KC. Plant protoplasts: Status and biotechnological perspectives. Biotechnology Advances 2005;23:131–71. https://doi.org/10.1016/j.biotechadv.2004.09.008. [8] Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, et al. Cells of the adult human heart. Nature 2020:1–10. https://doi.org/10.1038/s41586-020-2797-4. [9] Ma Q, Bücking H, Hernandez JLG, Subramanian S. Single-cell RNA sequencing of plant-associated bacterial communities. Frontiers in Microbiology 2019;10:2452. https://doi.org/10.3389/fmicb.2019.02452. [10] Rich-Griffin C, Stechemesser A, Finch J, Lucas E, Ott S, Schäfer P. Single-Cell Transcriptomics: A High-Resolution Avenue for Plant Functional Genomics. Trends in Plant Science 2020;25:186–97. https://doi.org/10.1016/j.tplants.2019.10.008. [11] Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, et al. Eleven grand challenges in single-cell data science. Genome Biology 2020;21:53. https://doi.org/10.1186/s13059-020-1926-6. [12] Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nature Protocols 2018;13:599–604. https://doi.org/10.1038/nprot.2017.149. [13] Xu G, Liu Y, Li H, Liu L, Zhang S, Zhang Z. Dissecting the human immune system with single cell RNA sequencing technology. Journal of Leukocyte Biology 2020;107:613–23. https://doi.org/10.1002/JLB.5MR1019-179R. [14] Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proceedings of the National Academy of Sciences 2020. https://doi.org/10.1073/PNAS.1919893117. [15] Wilbrey-Clark A, Roberts K, Teichmann SA. Cell Atlas technologies and insights into tissue architecture. Biochemical Journal 2020;477:1427–42. https://doi.org/10.1042/BCJ20190341.