Single cell RNA Sequencing (scRNA-Seq) is a powerful technique for deciphering the different cell types within a tissue sample. It gives an unprecedented insight into complex gene expression dynamics on a single cell basis, allowing researchers to understand biological systems on completely new level.
A number of parameters are paramount to making the most out of your single cell or single nucleusi data. To help you run successful scRNA-Seq and sNuc-Seq experiments, we have collated our top tips for single cell experiments below.
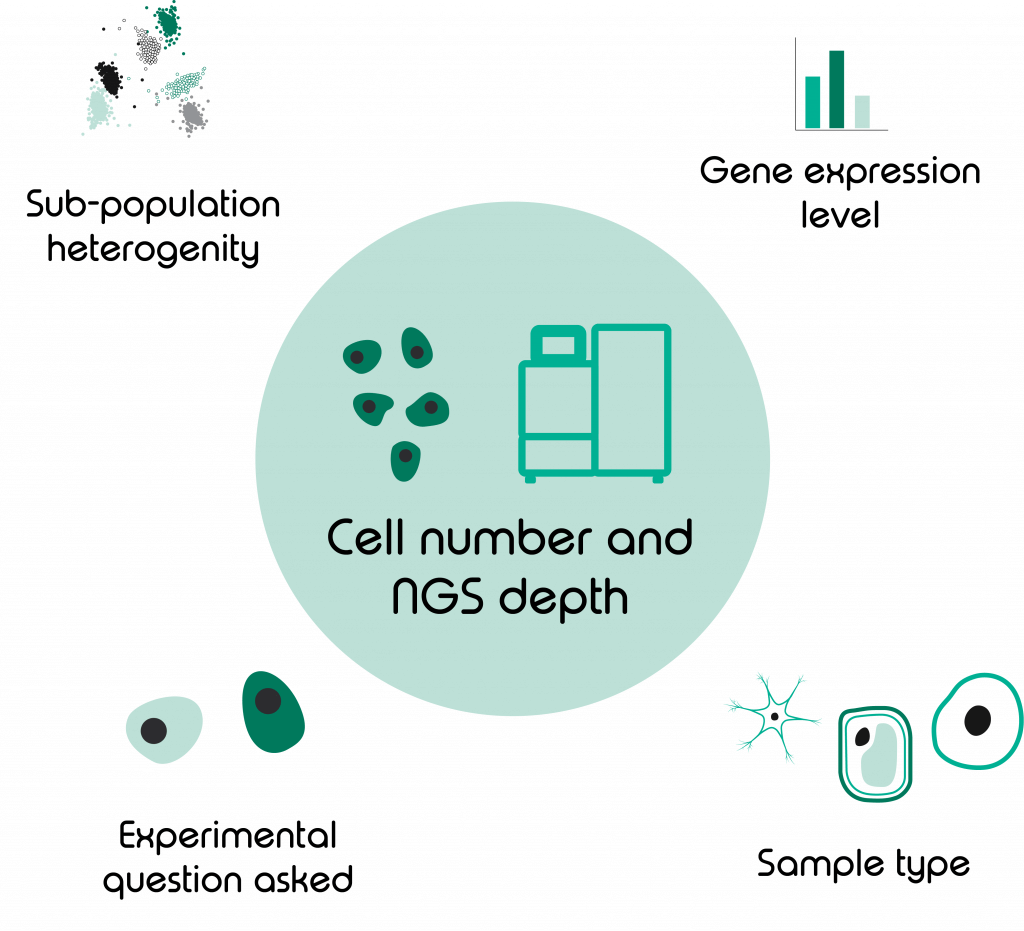
Choosing the right number of samples, cells, and sequencing depth for your experiment
Our customers often ask, “How many cells do I need to analyse?” or “How deep do I need to sequence?” The answer we give is not as easy as one might think and, unfortunately, it must be “it depends”.
More exactly, it depends on:
- Experimental question asked
- Sample type
- Level of sub-population heterogeneity
- Expression level of differentially regulated genes
Let me elaborate on those points; in a bulk RNA sequencing experiment, one is limited only by sequencing depth. In scRNA-Seq you will need to ask yourself if it is more important to sequence a high number of cells or to sequence deep. Then, unless you have unlimited funds available, it’s likely to be one or the other.
Is the aim of your project to detect as many different cell groups as possible and do you have a high level of sub-population heterogeneity? Then the answer probably is, analyse a higher number of cells. If a sample is heterogeneous, it is especially important to sequence enough cells to observe all representative cell types. The sequencing saturation per cell might be lower here but it will likely be sufficient to detect your sub-populations of interest. A nice tool to calculate how many cells you might need to analyse depending on minimum fraction of rarest cell type and desired minimum number of cells per cell type can be found on this page by the Satija lab.
Equally, there is a certain threshold of reads that is considered adequate to call a gene as ‘expressed’. Therefore, if your goal is to detect as many genes as possible you might want to sacrifice cell number for a higher NGS-depth. This will achieve a higher sequencing saturation per cell.
Besides this, the sample type will also determine if your emphasis should be on read or cell number. Some cell types due to high numbers of mitochondrial RNA or high cellular RNA levels require more reads per cell than others.
As a last point I also want to mention that if budget permits, run at least 2 replicates of your experiment. Dolomite Bio’s Nadia instrument will allow you to run up to 8 samples in parallel to reduce batch effects between runs.
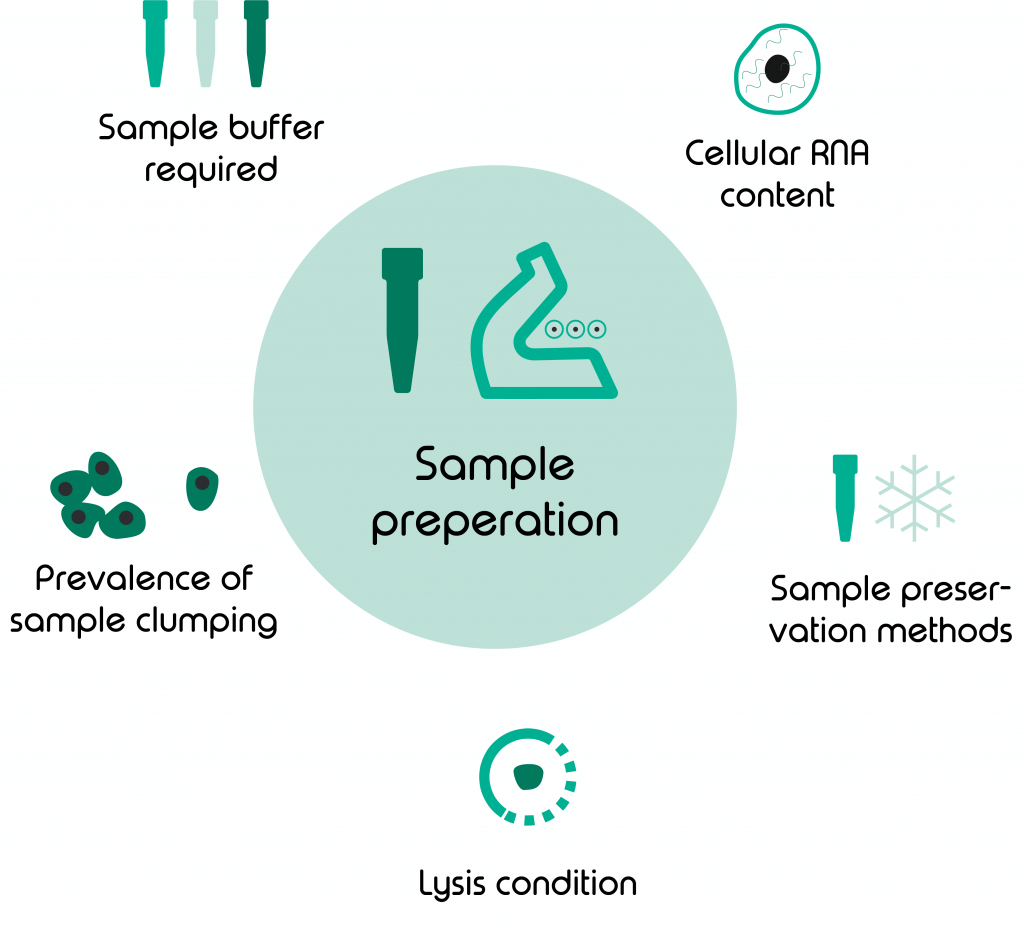
Sample preparation 101
Every sample or cell type comes with a specific set of requirements and limitations. To make the most of your sample its important to gather as much information about it and adjust your experiment accordingly. Here are some factors to look out for:
- Type of sample buffer
- Lysis conditions needed
- Prevalence of cells/ nuclei clumping
- Sample preservation methods
- Cellular RNA-content
Do you work with a standard mammalian sample that suspends well in PBS and lyses well just with Sarkosyl? That’s great! You will be able to use our standard protocol for scRNA-Seq on Nadia.
Now more than ever though, the field of single cell analysis is explored by researchers from a wide range of fields, working with species and samples that are more difficult to handle than the traditional mammalian samples. Often, those samples require different sample buffers to avoid clumping of cells or to ensure the viability of the sample.
This is particularly true when working with plant samples that can only be analysed when protoplasts have been extracted. Protoplasts, need special buffers that are quite a bit more viscouse than PBS and need careful adjustment of run conditions to successfully encapsulate sample and beads during a scRNA-Seq run.
This adjustment can be easily done with the Nadia Innovate, which allows for change of pressure, temperature, run time and stirrer speed. The latter parameter is particularly useful for those cell and sample types, such as nuclei, that are prone to clumping if left too long unsuspended in solution. Increasing stirrer speed and ensuring you work swiftly through your protocol will help reduce the occurrence of doublets.
Most often nuclei are used for samples where cells can not be lysed either enzymatically or chemically or for samples that have undergone preservation (e.g. through freezing). To give you an example, Storage of samples is often required for biopsies taken from patients. Although it is great that we can successfully store these samples for later use, the process of preservation somewhat renders cells membranes more fragile. This makes cells more prone to leaking RNA leading to cross- contamination between transcriptomes. To avoid such contamination during an scRNA-Seq run we typically recommend switching to our sNuc-Seq solution that works with single nuclei instead of cells.
With regards to RNA content, especially when planning your first single cell experiment on a sample type you haven’t previously worked with, it’s worthwhile checking what level of RNA content you should expect. This will greatly aid you in determining the number of PCR cycles required to amplify your cDNA library.
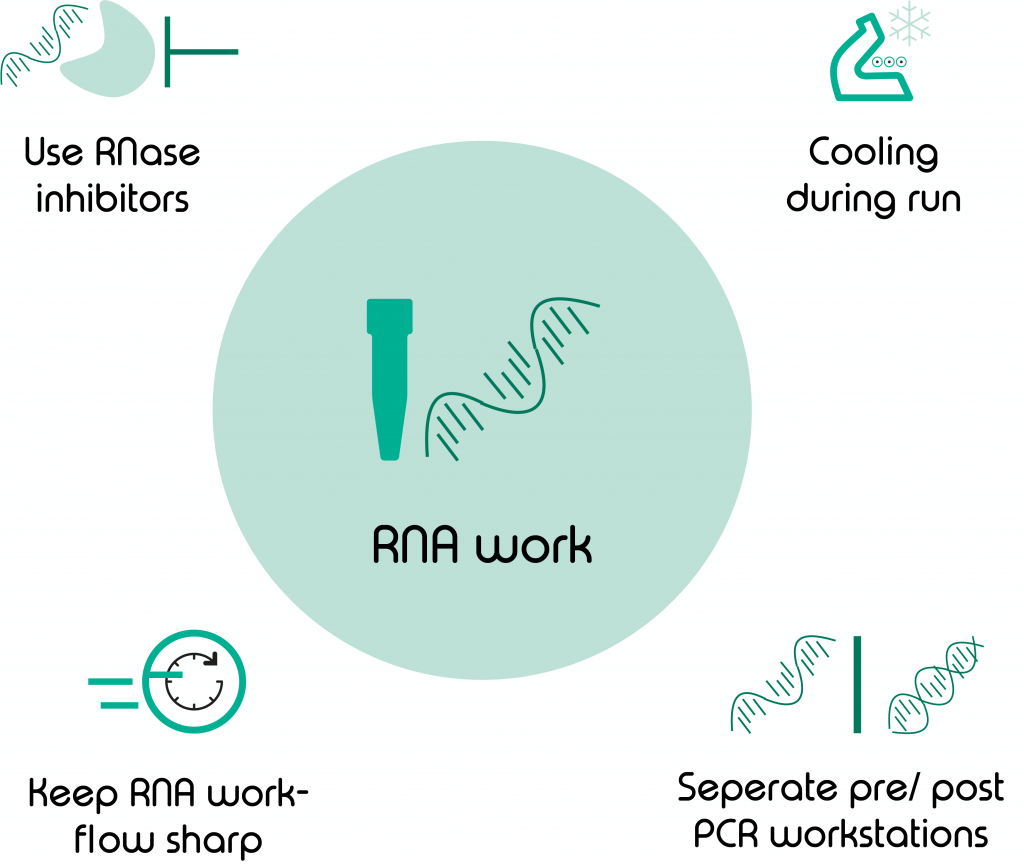
4 tips for good RNA lab techniques
The key point that you should always remember when performing your single cell-on Nadia- experiment is that you are working with RNA; as opposed to DNA, RNA is more fragile and prone to degradation by RNases. Therefore, I would like to give you these 4 simple rules to follow for each of your scRNA-Seq , sNuc-Seq and ppRNA-Seq experiments:
- Cooling during sample encapsulation
- Keep your RNA workflow swift and streamlined
- Use RNase inhibitors
- Keep separate pre- and post-PCR workspaces
Elevated temperatures lead to RNA degradation and can alter the cellular transcriptome during cell lysis and mRNA capture. Continued cooling during Nadia runs ensures minimal batch effects in between runs.
For any RNA work that happens outside of the Nadia, before as well as after the run, we recommend keeping your work swift and streamlined to avoid excessive RNA degradation and unwanted transcriptomic changes. Avoid long waiting times, especially when starting your scRNA-Seq experiment. This will help prevent clumping of cells, reduce cell death and premature lysis.
Furthermore, the use of RNase inhibitors will help you maximise data quality for your experiment.
Lastly, we have found it particularly important in our labs to keep separate pre- and post-PCR workspaces. Although, I appreciate this might not be possible in every lab, it is highly recommended if you can. This will greatly decrease the risk of amplicon or environmental contamination, which is particularly important for methods such as scRNA-Seq where RNA input is already a lot lower than in other methods.
So, this is it, if you follow the tips and tricks mentioned in this article you are sure to have successful single cell experiments in the future.
Further reading
If you would like to learn more about data quality, read this blog:
Or you can learn more about the handling of mRNA-capture beads here