Transforming Drop-Seq – scRNA-Seq on Nadia
In the fast-evolving world of genomics, a large amount of work has gone into helping Drop-Seq retain its place at the forefront of high throughput techniques.
At Dolomite Bio we have developed a single-cell RNA sequencing (scRNA-Seq) protocol that overcomes some of the challenges surrounding the original Drop-Seq technique.
Bulk RNA sequencing methodologies prevent the determination of individual cell types present in a sample as they provide an average read-out. However, there is a growing need to better understand sample complexity. Techniques, such as Drop-Seq, open many research possibilities as it permits to study the heterogeneity in cell populations at the single-cell resolution.
In 2015, Macosko et al. described Drop-Seq, a droplet-based microfluidic approach allowing the study of single-cell transcriptomes (Macosko et al. 2015). Using a microfluidic device, single cells are co-encapsulated with individual beads barcoded with oligos in droplets. Following encapsulation, cell lysis occurs inside the droplets and the mRNA is then captured on the beads. After emulsion breakage, reverse transcription occurs in bulk and the cDNA is amplified, resulting in libraries, which can then be prepared for sequencing.
1: Improved cell capture rate – a key parameter in Drop-Seq like approaches
The capture rate is a measure of the percentage of the initial single cells that will be sequenced. It corresponds to the percentage of cells that are co-encapsulated with a bead in the droplets. In other terms, the capture rate percentage is equal to the percentage of droplets containing a bead.
The capture rate of Drop-Seq and Drop-Seq-like approaches is not influenced by the technique or the instrument used to achieve encapsulation. The capture rate is, however, directly dependent on the initial concentration of beads, as a higher initial bead concentration permits a higher capture rate. Therefore, the type of beads, which can be hard and non-deformable or soft and deformable, plays an essential role as different bead types can be loaded at different initial concentrations (Figure 1).
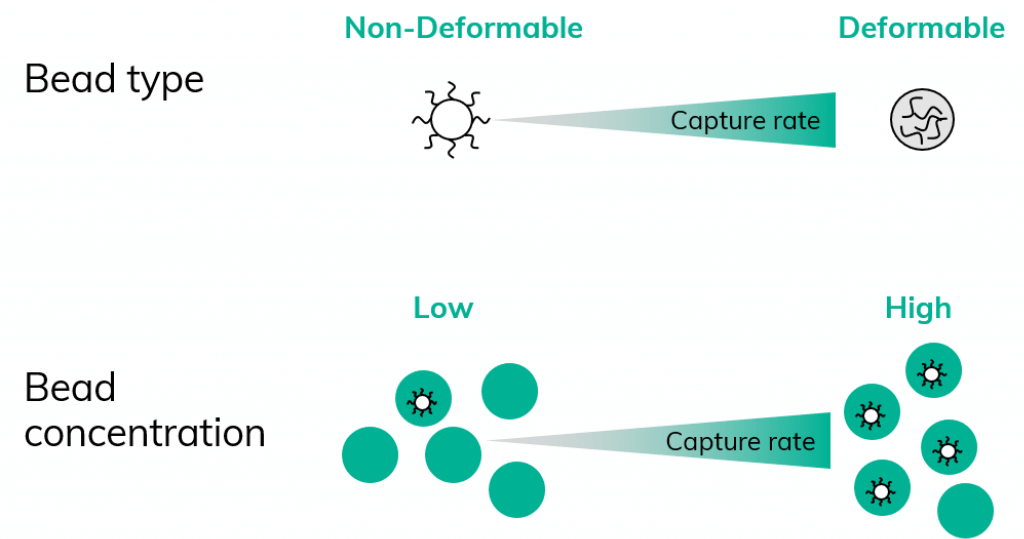
Chemgenes beads, which are hard non-deformable beads, are used in the scRNA-Seq on Nadia protocol at an initial concentration of 600 beads per µl. This initial bead concentration corresponds to a cell capture rate of approximately 11%. The initial hard bead concentration in this protocol was chosen to ensure a low bead doublet rate.
The Nadia Innovate provides a lot of flexibility, allowing modification of parameters and reagents and offering a unique possibility for scientists to develop protocols tailored for their own projects.
For example, deformable beads made of hydrogel can be loaded at much higher concentrations. Using the Nadia Innovate, scRNA-Seq can be performed and updated to achieve capture rates in excess of 70%.
2: Low cell doublet rate with the Nadia platform’s unique features
The average occupancy of droplets with cells depends on the initial cell concentration and on the volume of droplets that are produced. Increasing either the initial cell concentration or the volume of droplets produced would directly increase the average occupancy of droplets with cells. In microfluidic devices such as the Nadia platform, cell encapsulation in droplets follows a Poisson distribution. It is, therefore, possible to determine the number of droplets containing 0, 1, 2 or more cells. Increasing the volume of droplets or the starting cell concentration results in a higher number of droplets containing two cells or more.
However, there are other key parameters to consider. In most microfluidic platforms, cells are held in a reservoir for the duration of the run without a method for resuspension. Therefore, cells will gradually settle throughout the run, thereby inflating the doublet rate. To reduce the number of doublets and achieve truly single-cell data the Nadia instrument uses proprietary stirrer technology to ensure a homogenous suspension of cells during the entire encapsulation process (Figure 2).
In addition, the precision pressure control on the Nadia instrument generates highly monodispersed emulsions with low size variation ensuring a low doublet rate. Overall, these advantages allow scRNA-Seq on Nadia to retain low cell doublet rates even when assaying cell types or nuclei prone to clumping (Figure 2).
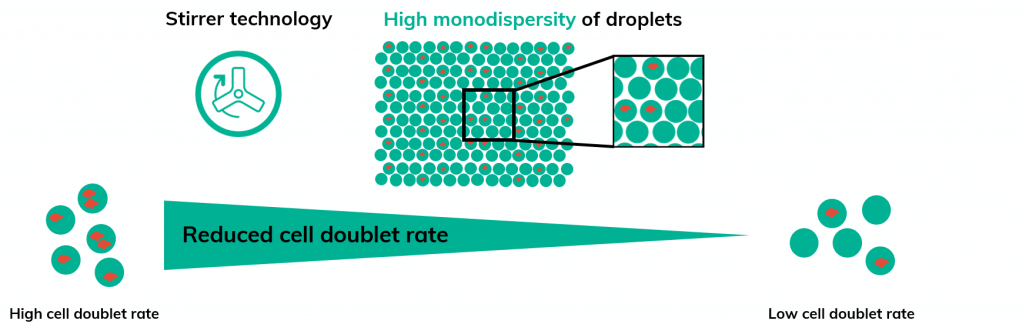
To assess the doublet rate, it is important to perform mixed species scRNA-Seq experiments. By mixing cells from two different species in a 1:1 ratio it is then possible to determine the number doublets containing more than one cell. Using both human HEK and mouse 3T3 cells, scRNA-Seq experiments on Nadia were conducted. The proportion of mouse to human mRNA molecules bound to each bead indicates where two cells from two different species are encapsulated with one bead. To account for same species doublets, the value corresponding to mixed species events must be multiplied by two.
It was shown, in mixed species experiment using scRNA-Seq on Nadia, that the cell doublet rate is between 4 and 7% at about 6,000 cells captured. It is important to note that this percentage can be reduced drastically if less cells are captured. For example, if capturing 3,000 cells you can expect a doublet rate of about 1.5 to 3%.
3: Perform more projects within budget Using the Nadia platform
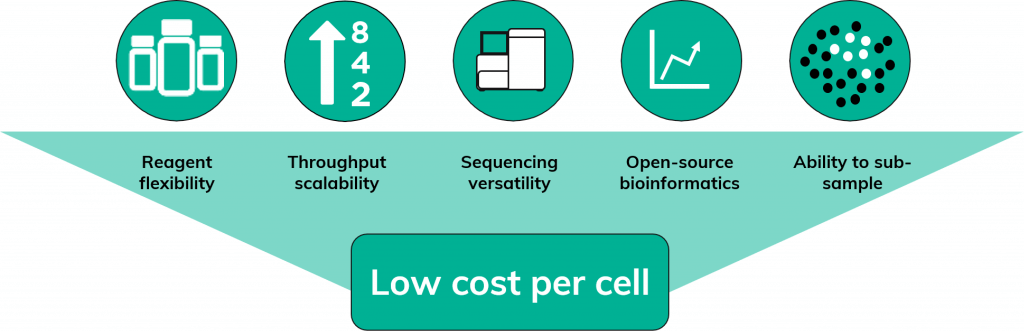
Single-cell transcriptomic workflows can be expensive, especially when they are bound to exclusively commercially available reagent kits. Nadia users can either purchase fully quality-controlled kits from Dolomite Bio or buy reagents from suppliers. As our protocols are open source, we provide a detailed list of reagents with their part numbers. This allows users to purchase reagents directly from suppliers. By doing so, it is possible to performed scRNA-Seq on Nadia at a fraction of the cost of other scRNA-Seq kits. This reagent flexibility is just one of several points that count towards the affordability of scRNA-Seq on Nadia, allowing researchers to complete more projects within budget.
scRNA-Seq also benefits from throughput scalability when run on the Nadia instrument. 1, 2, 4 or 8 samples can be run simultaneously, each capturing up to 6,000 single-cell transcriptomes in a 20-minute run. In addition, this scalability comes without any significant cost implication, giving users the possibility to choose the cartridge that best fits their experimental needs.
The cDNA libraries produced using the Nadia protocols are compatible with all the sequencing platforms available on the market. Moreover, the bioinformatic analysis can be conducted using a fully open source bioinformatic pipeline such as dropSeqPipe.
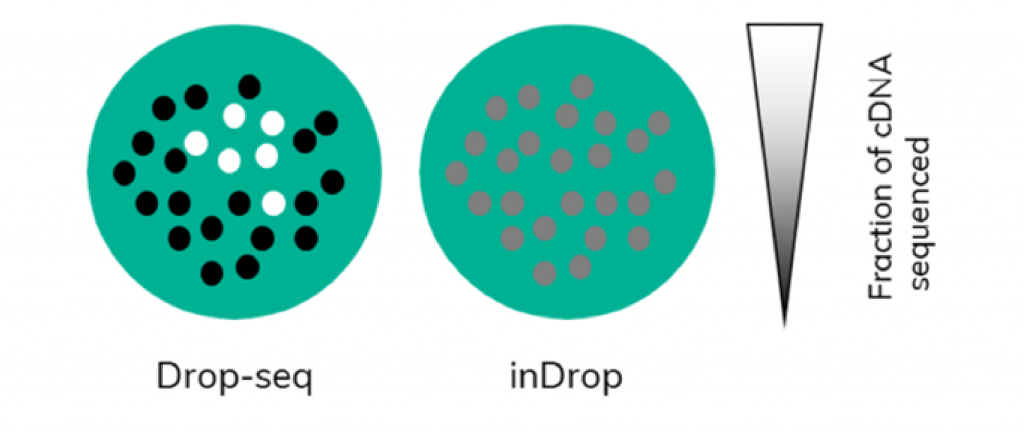
Additionally, the workflow of scRNA-Seq on Nadia uniquely allows for subsampling of all the transcriptomes harvested from a single sample as the amplification step is carried out on cDNA that is still bound to beads. In Drop-Seq, following RT, which is done in bulk on all beads, PCR amplification starts from bead-bound cDNA. Users can choose the number of cells they wish to sequence by picking a certain number of beads to undergo PCR. This will enable them to retain information about complexity because amplification happens on the entirety of a single cell transcriptome (Figure 3). This allows the user to test a representative subset of cells to gain insight into expression levels of the full sample.
However, it not as straightforward to conduct subsampling in other methodologies such as when using inDrop. In inDrop, the cDNA is released from the beads inside the droplets following RT. When the emulsion is broken to recover the cDNA, all the captured single-cell transcriptomes are mixed and amplification is carried out on mixed cDNA. Subsampling at this point would equate to taking a small amount of cDNA originating from each captured cell rather than full transcriptomes. This potentially biases the data towards genes with higher expression levels, as only a fraction of the total cDNA of each cell would be analysed (Figure 3). More information about Drop-Seq and inDrop methodologies can be found here.
Each of these measures add up to make scRNA-Seq on Nadia a highly affordable form of, not just Drop-Seq, but of single-cell sequencing in general (Figure 4).
4: scRNA-Seq on Nadia is fast and convenient
From its inception in 2015, Drop-Seq has undergone many optimisations. At Dolomite Bio, we have streamlined the Drop-Seq process to improve ease of use and decrease the time spent at the bench.
First, scRNA-Seq on the Nadia instrument is highly scalable; up to eight samples can be run simultaneously, each generating up to 6,000 single-cell transcriptomes in a 20-minute run. The protocol uses a rapid filter-based emulsion breakage method which is unique to scRNA-Seq on Nadia. Finally, the reagent kit for scRNA-Seq on Nadia does not require buffer preparation, which allows for fast library generation.
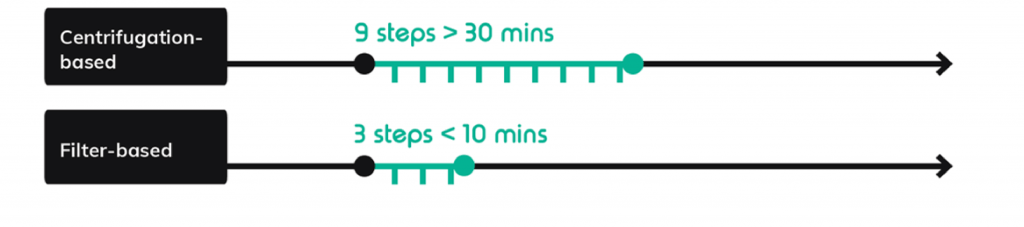
Instead of more complicated chemical emulsion breakage techniques common to some droplet based-workflows, a filter-based emulsion breakage step is used in the scRNA-Seq on Nadia protocol. It decreases the number of discrete steps from nine to three and decreases hands-on time from 30 minutes to less than 10 minutes per sample (Figure 5).
The data yield is improved by decreasing the number of washing steps and the time taken to proceed to cDNA. Using scRNA-Seq on Nadia, it is possible to retain more beads, obtain more STAMPs and reduce the impact of mRNA degradation.
With the time saving measures integrated into the scRNA-Seq on Nadia protocol, sequencable libraries can be obtained from singulated cells comfortably within one and a half days.
Summary
The scRNA-Seq protocol developed by Dolomite Bio, overcomes some of the challenges surrounding the original Drop-Seq technique. Soft and hard beads can be used on the Nadia instrument and the Nadia innovate allowing users to choose the most appropriate bead type based on their experimental needs and the cell capture rate required. Dolomite Bio’s platforms present several unique features compared to other commercially available microfluidic devices such as stirrers, a temperature controller and three independent pressure pumps. The stirrers ensure that both the cells and the beads remain well homogenised, reducing doublet rates.
Moreover, the scRNA-Seq protocol on Nadia is open source allowing for reagent flexibility. This, combined with throughput scalability, sequencing versatility, free bioinformatic pipelines and the possibility to subsample, make scRNA-Seq on Nadia an affordable form of scRNA-Seq; allowing you to perform more projects within budget.
Finally, scRNA-Seq on Nadia was developed to be fast and easy to perform. The protocol has been fully optimised with, for example, the addition of a filter-based emulsion breakage step. This step, by reducing the time taken to proceed to cDNA, leads to improved data yield as samples are less affected by mRNA degradation.
References:
Macosko, Evan Z., Anindita Basu, Rahul Satija, James Nemesh, Karthik Shekhar, Melissa Goldman, Itay Tirosh, et al. 2015. ‘Highly Parallel Genome-Wide Expression Profiling of Individual Cells Using Nanoliter Droplets’. Cell 161 (5): 1202–14. https://doi.org/10.1016/j.cell.2015.05.002.#
Further Reading
Interested in learning more about Drop-Seq? We have created some other blogs on the topic for you:
The benefits of Drop-seq-like approaches for single cell sequencing


