Getting started with single cell research
In the last few years single cell research has developed into one of the most successful emerging fields in clinical and academic research. Since the establishment of single cell RNA-sequencing (scRNA-Seq) over 10 years ago (Tang et al. 2015), a plethora of new single cell applications and technologies have made their way into our labs.
Between all the different techniques and commercial instruments that are now available, it can be hard to decide where to get started if you are new to the field.
But don’t fret, we are here to help you!
In this article I will summarize the why, what and how to perform single cell research to make it easier for you to get started.
First things first, a short introduction to bead-based scRNA-Seq methodologies. If you already know how the technology works, feel free to skip this intro.
Single cell RNA-Seq 101
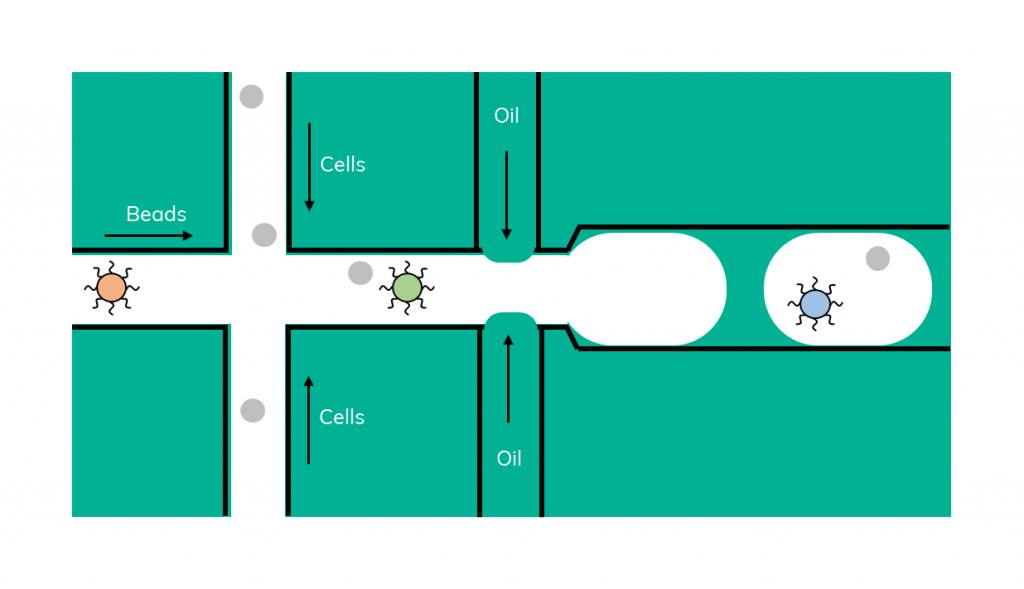
In 2015, Macosko et al described Drop-Seq, a droplet-based microfluidic approach allowing the study of single cell transcriptomes (Macosko et al. 2015).
Using a microfluidic device, single cells are co-encapsulated with individual oligo-coated barcoded beads in droplets. Following encapsulation, cell lysis occurs inside the droplets and the mRNA is then captured on the beads. After emulsion breakage, reverse transcription occurs in bulk and the cDNA is amplified, resulting in libraries, which can then be prepared for sequencing.
Which biological questions can be answered through single cell research?
The power of scRNA-Seq is that you can look at the individual transcriptomes of thousands of single cells within a given sample at the same time. This fact alone has got many people excited in the field and led to a large number of publications about single cell sequencing.
However, which biological questions do single cell methods really answer? We have summarized some of the most important ones for you below:
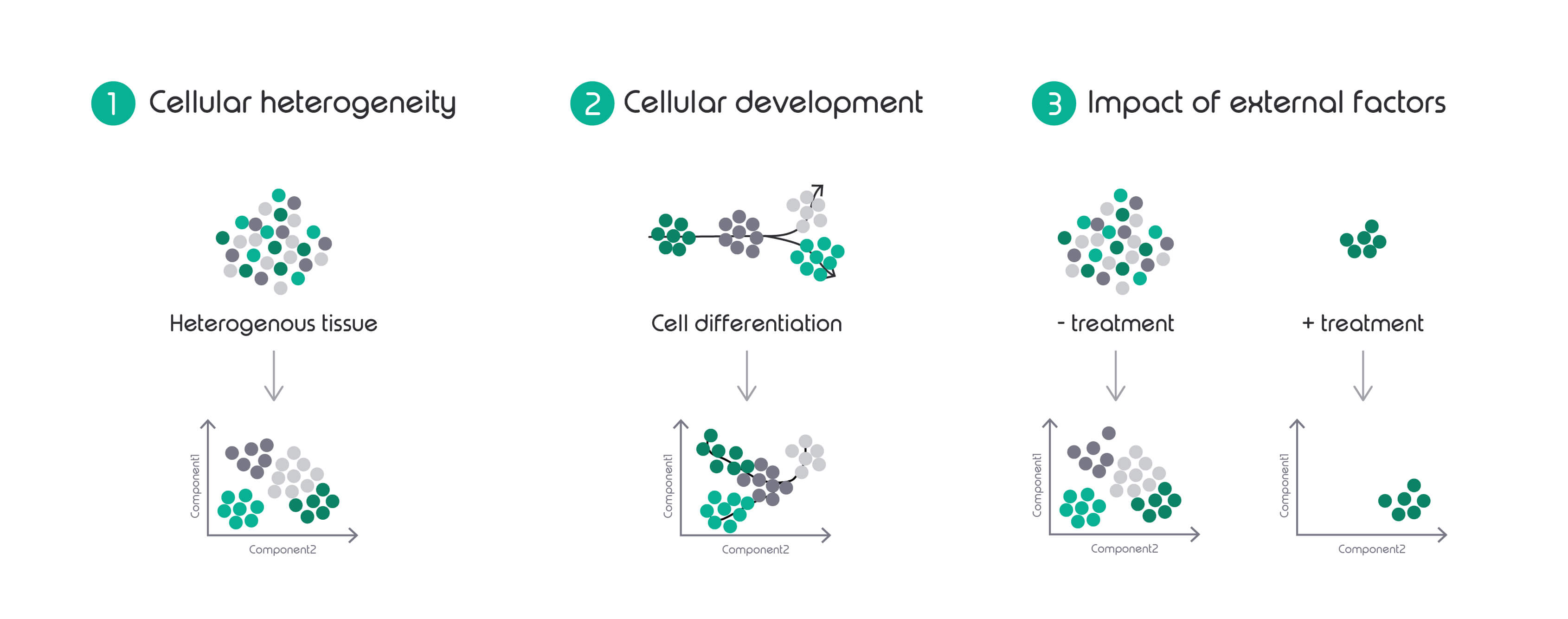
1. Cellular heterogeneity
2. Cellular development
3. Impact of external factors
The first biological question answered by single cell workflows that most people can think of is cellular heterogeneity. This looks at the number of different cell clusters within a sample that have very similar expression levels and are, therefore, grouped together. This might help with the identification of different cell types present in an organ, a tissue or an organism.
Single cell research has the potential to be a lot more powerful than that; it can give insight into developmental processes and genes associated with them. This is called lineage tracing and it enables the construction of complex transcriptional atlases of adult tissues and developing embryos from the transcriptomes of individual cells.
Lastly, single cell techniques can support the study of external factors on tissues, organs or whole organisms. This can be immensely helpful, particularly when studying the effect of a treatment on cancer tissues. These often contain smaller sub-populations that might not be as susceptible to the treatment as the main populations of cells detected by bulk RNA-Seq methods.
Choosing a single cell technology that fits your project
The underlying technologies in single cell research can broadly be separated into 3 different types.
1. Plate-based
2. Nano & Micro well-based
3. Droplet-based
Which technology is right for you depends on sample type, number of cells available per sample and total number of samples to be processed per experiment.
The plate-based methods are the simplest and, as the name alludes to, are carried out on 96 or 384 well plates. Single cells are placed into individual wells, either manually or through automation. As you might have guessed, this technology is only suitable for very low numbers of cells and samples. However, if you have just a couple hundred cells in your sample, this method is perfect as a few hundred cells is too little of an input for any of the other technologies and it will allow you to save cost.
In contrast, micro well and droplet-based technologies are much better suited for large numbers of cells, but only droplet-based technologies provide high sample throughput as well (up to 8 samples in one run, or more if multiplexed).
Nano & micro well-based methods are similar to (macro)well-based methods in that they have wells in which individual cells are distributed. However, as the wells are very small and are sitting on a chip, cells are evenly distributed automatically by surface tension and settle in through gravity alone into the wells.
Droplet-based methods operate on microfluidic chips in which, by combining oil with a water phase, single aqueous droplets are created. Cells and beads are flowed into the chip in the water phase and encapsulated together in the droplets. Cells are lysed due to the presence of lysis buffer and their mRNA is captured onto the beads.
In summary, if you have very few cells to start with, use a well plate. If you have high cell and sample numbers, use droplet-based methods such as scRNA-Seq on Nadia.

What to consider when choosing a commercial platform
As more biological fields move into the realm of single cell genomics, personalized medicine and high throughput cellular screening or analysis, there has never been a better time to consider droplet-based single cell technology for your research.
But how can a researcher ensure that their equipment is both robust and future proof enough to meet potential advances in the rapidly advancing field of single cell genomics? Researchers, from clinical applications to basic research to applied industry, should take care to consider these must-haves when equipping their labs for the single cell revolution:
• Low doublet rates – Sample stirring to avoid settling or clumping of samples.
• Broad sample compatibility –Suitability for a wide range of tissue types and organisms.
• Application versatility –Option for protocol development.
• Exceptional data yield – High gene capture rate even at low sequencing depths.

Simplistically, the central process within many single cell droplet-based workflows is the distribution of cells and oligo capture beads into droplets. This can be extremely efficient, but if it is not tightly controlled, multiple cells can end up in the same droplet, creating doublets in the resulting data. High doublet rates must be avoided. Why invest in a single cell platform if it does not yield data from true single cells?
Equally important is the compatibility of a system with a wide variety of cell types. Differences in morphology and physical size of cells can impact whether these cells can flow unhindered through a microfluidic system, potentially introducing biases in the resulting data. The ability to alter buffer composition and droplet size can allow improved flexibility for cell types large and small, like bacteria or nuclei (Davey et al. 2020), or cells that require specialized buffer conditions, such as plant protoplasts (Davey et al. 2005). Illustrating this, cardiomyocytes are major cells of interest in prospective cell atlases, but their large size prevents them from being dropletized whole by some microfluidic systems. By contrast, single cell platforms like the Nadia instrument can encapsulate cells of over 50µm in size, broadening the range of cell types accessible for transcriptomics.
Any single cell platform worth its salt must, therefore, be sufficiently open to perform new and potentially unforeseen applications. Whilst locked-in systems with a discrete set of applications are typically popular for their ergonomic design, users can be left behind when ground-breaking new molecular techniques surface. With a system capable of open protocol development and optimization, such as the Nadia Innovate, researchers can proactively develop applications to take advantage of the latest advances in sequencing technologies.
Lastly, applications like scRNA-Seg prioritize sequencing metrics. One of the main metrics is gene capture rate. This is a term for how many genes one can expect to capture from each single cell at a given sequencing depth. When comparing datasets from different single cell platforms, it is important to standardize this data according to both sequencing depth per cell and throughput.
How to get started in single cell research
Now that you have learned the theory of what system might be suitable for your sample and research, it is time to decide.
As easy as this might sound it is still a daunting task. We all know theory is not practice, especially when it comes to research where a million things might suddenly behave differently when you actually perform your experiments in the lab.
More often than not, you might not need to purchase a full system but just need 1 or 2 single cell experiments to complete your research for a paper.
Additionally, investment in a commercial single cell system is capital spending and often requires grant application or at least a tender. So, in some cases, it might be nice to “try before you buy”.
Most of these issues can be resolved by looking into using a Single Cell Consultancy Service that enables you to try a certain scRNA-Seq platform, test the compatibility with your sample type and gain the experimental data to complete a grant application or submit that research paper.
At Dolomite Bio we offer a specialist service for single cell and single nucleus RNA-Seq that can help you get started. We offer personal support by one of our single cell specialists, who have a combined over 15 years of experience in the field. We are able to run your samples through our workflow from cell isolation all the way to data analysis by our in-house bioinformaticians. Importantly, you can pick and choose which steps you would like us to perform and which not to perform for cost effectiveness.
Would you like to learn more about our Single Cell Consultancy Service? Join us at our next webinar here!
Further Reading
Enjoyed this blog post? You can read more about scRNA-Seq on Nadia vs Drop Seq
Top 4 Improvements in Drop-Seq for scRNA-Seq research on Nadia
And you can learn more about the possibilities available to you with the Nadia Innovate’s custom parameters.
Customizing single cell research protocols with Nadia Innovate
Or discover the most important things consider for your single cell platform

